|
|
Welcome to Yeast KID, the kinase interaction database. Yeast KID may seem daunting at first, but general use of the database should be fairly straightforward.
How do I search for interactions with a specific kinase or gene?
How do I search for genes which have interaction with more than one kinase or vice-versa?
How do I search for specific publications?
How do I search for high-confidence interactions?
How do I use the color filter feature?
How do I obtain the data?
How do I visualize the interaction network?
How do I search for interactions with a specific kinase or gene?
Searching for interactions with a specific gene or kinase is possible through the input boxes. Yeast KID allows the search of all verified ORFs and 127 kinases through different nomenclatures. These include the systematic name (eg. YDR146C), the gene name (eg. SWI5), or the SGD:ID (eg. S000002553). We recommend using the systematic name.
Multiple genes or kinases can be searched simulteanously by separating the queries by a space, a comma, or a semi-colon (eg. SWI5 MCM3 will search for both SWI5 and MCM3 at the same time). It is important to note that entering genes in both the gene and kinase fields will ask explicitly for the pair(s).
A checkmark will validate if the entered queries are present in our database. If not, it will indicate which entries are not recognized. Try it here!
How do I search for interactions with more than one kinase and vice-versa?
Yeast KID allows the user to search for overlapping kinases or genes. That is, given multiple genes as queries, Yeast KID can return a list of kinases which are shown to interact with the list of genes. The converse is possible, given a set of kinases, Yeast KID can return a list of genes phosphorylated by all the queried kinases.
To perform this type of search, input more than one kinase or gene in the input box and click on 'compute gene overlap' or 'compute kinase overlap'. Note that these buttons perform a normal search if there is not more than one gene or kinase in the input fields.
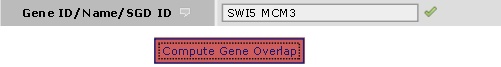 This will find all the kinases which interact with MCM3 and SWI5. This will find all the kinases which interact with MCM3 and SWI5.
How do I search for specific publications?
Interactions shown by a specific publication are queried by their PubmedID. As the gene and kinase field, Yeast KID allows the search of multiple PubmedIDs at the same time. Further, PubmedIDs can be exluded using an exclamation mark (!) before the PubmedID. Try it here!
How do I search for high-confidence interactions?
Yeast KID scores each interactions by its likelihood of being a gold-standard interaction. Therefore, interactions with a higher scores are more likely to be bona fide kinase-substrate relationships. The user can limit the interactions by a score threshold.
By default, the score is set to -5, which retrieves all the interactions in the database. Clicking on the corresponding help bubble shows the recommended score thresholds for statistically significant values.
How do I use the color filter feature?
The color filter feature allows for more complex queries with no special vocabularies. For example, most websites allow for these queries using combinations of parentheses and +/- signs. The color filter feature performs this vocabulary for the user.
The color filter feature is intuitive. The palette allows the user to 'paint' experimental categories of different colors. Painting a category, for example, HTP in vitro kinase assays in yellow, will retrieve all interactions shown by this category. Painting another category with the same color will allow interactions from either of these two categories.
Painting different categories using different colors will request interactions shown by both groups.
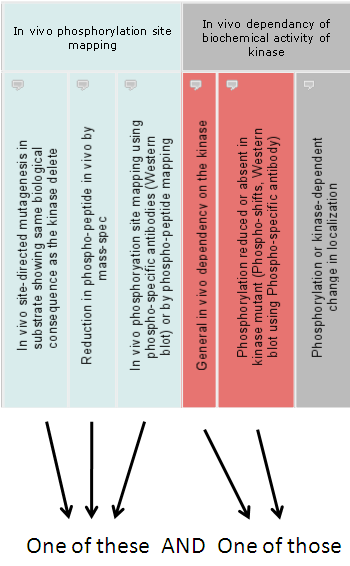
For further convenience, we allow two additional color classes, the inclusive green (+) will instead group the categories as 'All of these', where the interaction requires all the selected green experiments. This is similar to coloring each experiment of a different color.
We also have the exclusion white (-) which groups the category as 'None of these'.
How do I obtain the data?
All data queried from KID can be downloaded to a tab-delimited file. The tab-delimited file does not display checkmarks, but instead displays the specific notes and reference information for the interaction. Simply click the 'download table' link to obtain the data. The header of the tab-delimited file displays the specific query.
How do I visualize the interaction network?
For convenience, Yeast KID allows queries to be visualized in a Cytoscape-Web interactive network. These interactive networks can be downloaded to a Cytoscape compatible format (via import network function) or as a high-resolution image.
|
|