The ConDens Browser is a tool designed to facilitate the analysis of ConDens prediction. It can be used to load data generated by the ConDens Predictor program and to visualize the multiple sequence alignments of individuals proteins and motifs that are ranked by the predictor.
This browser is integrated with Jalview, which serves as the platform of alignment visualization, and is equipped with a library of motifs that permit convenient observation of a variety of motifs on the sequence alignments.
Overview
The ConDens Browser is composed of 4 windows:
This browser is integrated with Jalview, which serves as the platform of alignment visualization, and is equipped with a library of motifs that permit convenient observation of a variety of motifs on the sequence alignments.
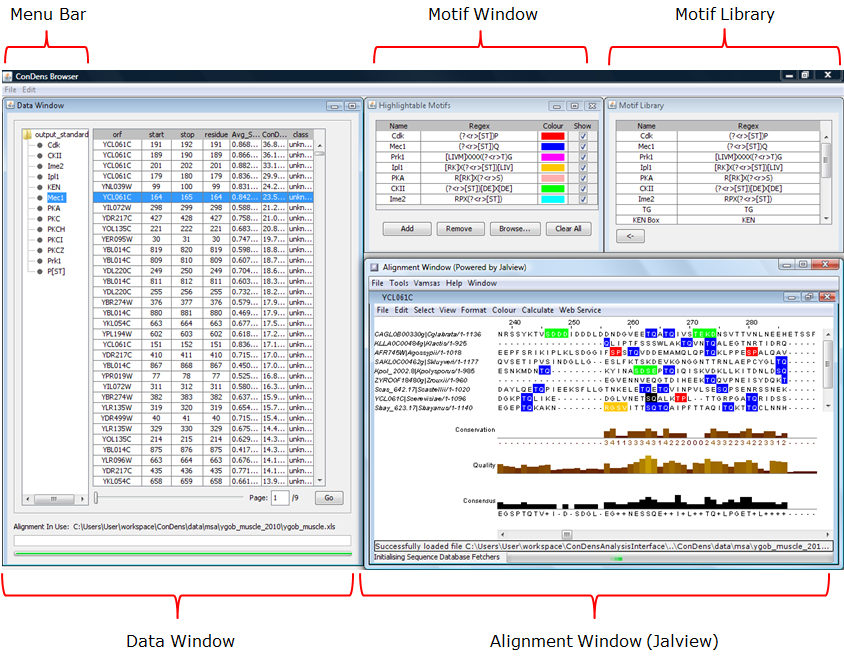
Figure 1: The ConDens Browser G.U.I. and sub-components. |
- Data Window: Displays ConDens prediction data in tabular form.
- Motif Window: A listing of motifs that can be highlighted on the Alignment Window
- Motif Library: A library of literature-derived motifs that can be added to the Consensus Window
- Alignment Window (Jalview): The Jalview alignment viewer that shows local alignments of individual data points
Data Window Motif Window and Library Alignment Window
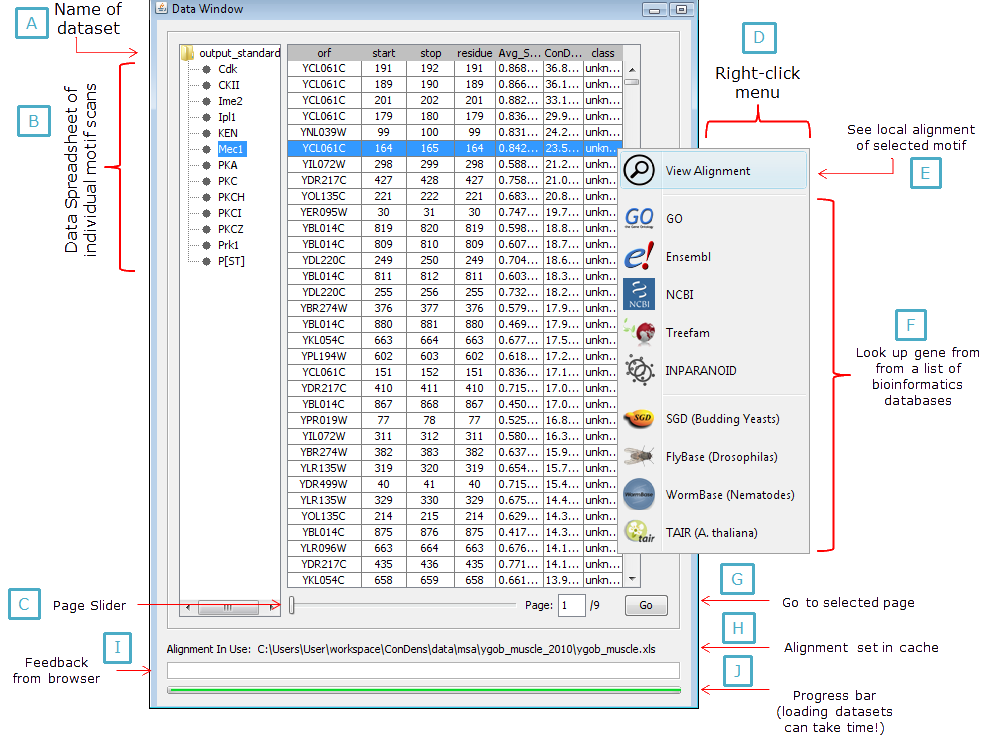
Figure 2: Data window and sub-components. |
This window (Figure 2) is designed to show tabular data generated from the ConDens Predictor program (specficially
for details). The dataset itself can be directly loaded onto the table using the menu bar (Figure 3). The file that the user needs to is a folder where he saved his ConDens prediction data to. Once an appropriate folder is selected, the program will look through all sub-folders inside the specified program for ConDens prediction data and then organize them into a file tree at the left of the window (Figure 2A-B). This process may take a while depending on the size of the datasets to be loaded and a progress bar (Figure 2J) is used to indicate how close the program is to finish loading the data input. For information on how the ConDens input data is structured, see Structure of ConDens Ouput Data.
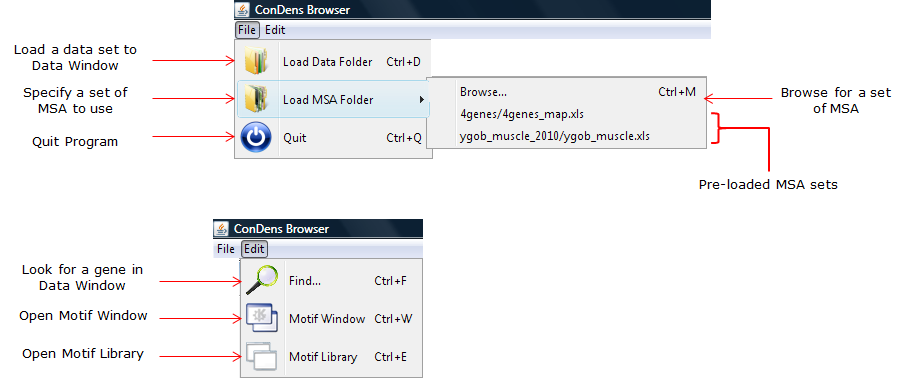
Figure 3: Menu bar. |
The individual data tables can be enormous in size - S. cerevisiae proteome alone has 4000+ proteins containing S/T-Q motifs. In order to accomodate the need of viewing large tables of data without overloading the user's cache, the browser divides up the table into "pages" that contain 2000 rows each, which are loaded on demand and off-loaded when not needed. The entirety of a data table can be navigated through a scroll bar (for intra-page surfing) and a page slider (for transiting to different pages, see Figure 2G). The mouse-wheel can also be used to scroll through pages (and across page boundaries) when Ctrl is held down.
Individual genes can be looked up in a particular data table using the Find... option in the menu bar (Figure 3).
Sorting the data
The data table as a whole can be sorted by a given column in ascending order or descending order by left-clicking or right-clicking (respectively) the column header. The sorting process can take a few seconds for large tables.
Viewing local alignments of a motif
The individual rows on the data table contain the coordinates of a motif and the name of its protein. To show the local multiple sequence alignment of the motif, the user can either do Ctrl + Left Click on the row of interest or right-click on the row and choose View Alignment from the pop-up menu (Figure 2E). Once that's done, the program will load up the appropriate sequence alignment in the Alignment Window and center at the appropriate coordinates (i.e. where the motif of interest is at). For information on browsing an alignment in the Alignment Window, see the sections below.
Looking up a gene
The proteins of individual motifs shown on the data table can be looked up. When the user right-clicks a row in the table, a pop-up menu will appear (Figure 2D) with a list of bioinformatics databases (Figure 2F). Choosing one among the list will open up a browser tab that directs the user to a page that corresponds to the protein of the row being selected. It is up to the user's discretion to choose an appropriate web database for his data - For example, a query on p53 in SGD (which is a Budding Yeast database) will not yield anything useful (as of October 2011).
Multiple Sequence Alignments
In order to display multiple sequence alignments for the input data, the user must specify a set of alignments to be used through the popup menu (Figure 3), which will then be indiciated in Figure 2H. Ideally, this set of alignment is same as the one used for the generation of the input dataset. If an alternate alignment set is to be used for whatever reason, the user must make sure it is properly structure and the nomenclature of the genes of interest are consistent (i.e. do not use UniProt names for the input data and then Ensembl names to label the aligned sequences). It is important to note that the ConDens Browser will display an alignment for a protein/motif of interest iff the information for the protein's alignment is available and properly formatted. For information on how the alignment data is structured, see Structure of Alignment Input.
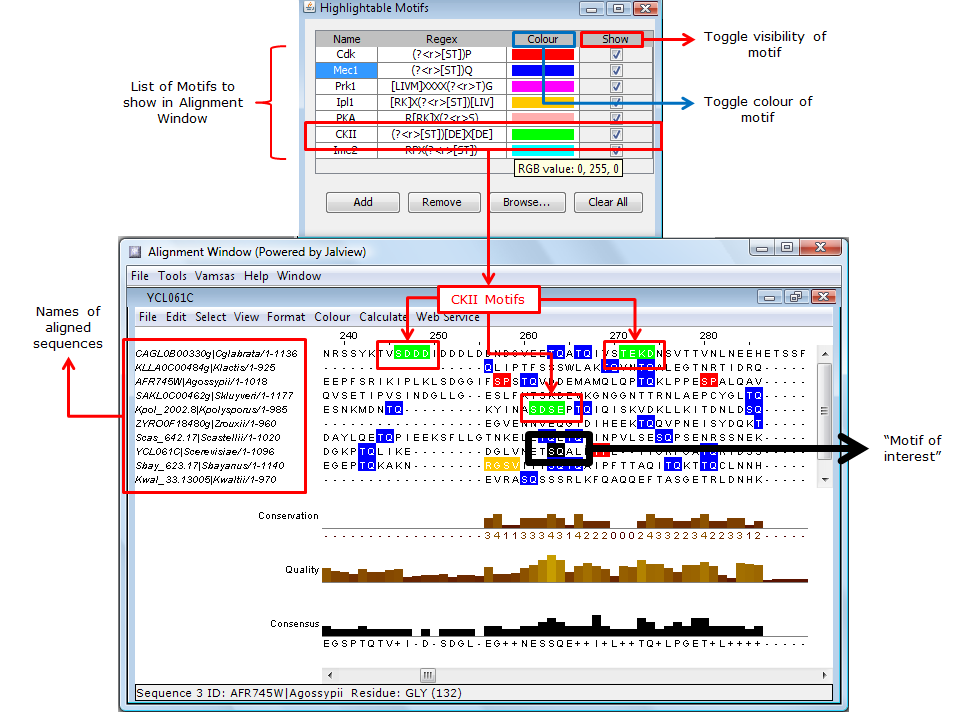
Figure 4: Hightlighting motifs in the sequence alignment. The motif window shows a list of motifs that can be hightlighted on a multiple sequence alignment displayed in the alignment window (Jalview). |
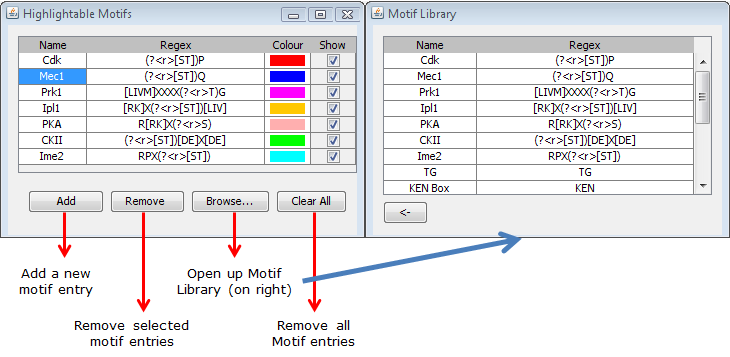
Figure 5: Functions of various buttons in the motif window. |
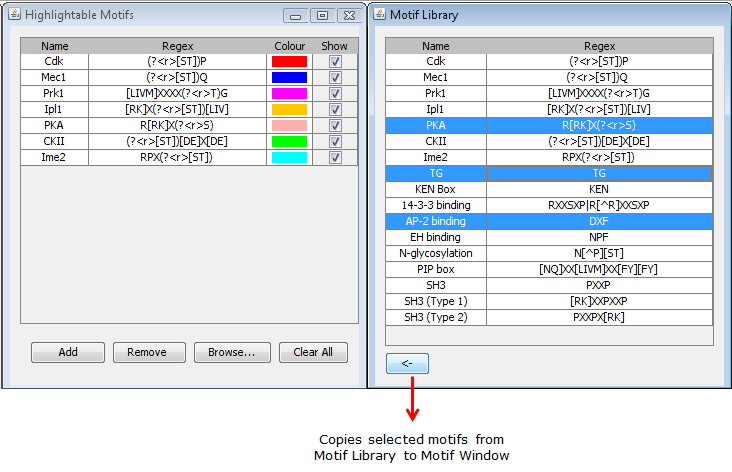
Figure 6: Adding more motifs to motif window from the motif library. |
The Browse... button on the window will open up the motif library (if closed), which contains a list of literature-derived motifs (which are too numerous to be conveniently included in this window). Motifs from this library can be copied over using a the <-- button (Figures 6). The entires of the motif library cannot be modified.
The Jalview alignment window is a professional software developed by Waterhouse et al. at the University of Dundee (website, paper). Documentation of its use can be found here.