Adding new presets
ConDens Predictor automatically looks for pre-available proteomes and alignments sets in the local directory. A new proteome or alignment set can be added by placing the relevant data in a new folder under
data/proteins or
data/msa, respectively. This new folder should contain a file called
info.xml that has the following structure:
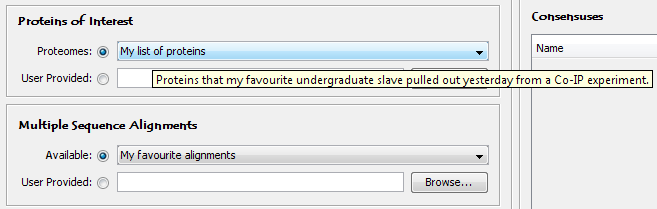
Sample info.xml file
<settings>
<proteins>
<name>My list of proteins</name>
<description>Proteins that my favourite undergraduate slave pulled out yesterday from a Co-IP experiment.</description>
<path>gene_list.txt</path>
</proteins>
</settings>
|
Default Settings of ConDens Predictor
It can be very bothersome having to re-pick input parameters and type up the same motif regular expressions over and over again when one opens the program each time. To reduce this annoyance factor, preferred input settings of this program can be specified in
data/default.xml. This file takes the exact same form as the command-line XML input of the program. See
here.
Default Settings of ConDens Browser
The default settings of ConDens Browser can be defined in
data/default_viewer_settings.xml. This file has almost the exact same structure as
data/default.xml but with fewer nodes:
Sample default_viewer_settings.xml
<settings>
<param rowlimit="2000" />
<output path="yeast_output_data" />
<msa path="msa/my_alignments/mapping_file.tab" />
<consensuses>
<consensus name="Cdk" regex="(?<r>[ST])P" />
<consensus name="Mec1" regex="(?<r>[ST])Q" />
<consensus name="Prk1" regex="[LIVM]XXXX(?<r>T)G" />
<consensus name="Ipl1" regex="[RK]X(?<r>[ST])[LIV]" />
<consensus name="PKA" regex="R[RK]X(?<r>S)" />
<consensus name="CKII" regex="(?<r>[ST])[DE]X[DE]" />
<consensus name="Ime2" regex="RPX(?<r>[ST])" />
</consensuses>
</settings>
|
Changing the Library of Motifs
The repository of short linear motifs provided are not very comprehensive. Should there be a need to make changes to it, the relevant file to modify is
data/consensus_library.xml, which stores a list of motifs and their information. This file has the following structure:
Sample consensus_library.xml
<library><consensuses>
<consensus name="Cdk">
<regex>[ST]P</regex>
<ref>
<text>Hunter T, Plowman GD: <b>The protein kinases of budding yeast: six score and more</b>. <i>Trends Biochem. Sci </i>1997, <b>22</b>:18-22.
</text>
<id type="PMID">9020587</id>
<id type="URL">http://www.ncbi.nlm.nih.gov/pubmed?term=9020587</id>
</ref>
</ref>
...
</ref>
</consensus>
<consensus>
...
</consensus>
</consensuses></library>
|