The output of ConDens Predictor is always exported to a folder specified by the user. This folder, if not modified, should contain a list of sub-folders and one for each of the motifs scanned by the program. For example, if an user runs the program on motifs named "Cdc2", "Rad53", and "NES Class 1a", then he should be able to find folders of the same name inside their specified output folder for the program-run (Figure 1).
Each motif folder contains 2 tab-delimited files: site_results.xls and protein_results.xls . The former contains the prediction scores and coordinates for each individual motif match and the latter contains the prediction scores for each protein (which are simply the maximum prediction score calculated for motifs in the protein).
There are 7 columns in eachsite_results.xls file:
There are less columns inprotein_results.xls files. Namely, only orf, ConDens_Score, and label remain.
Comments:
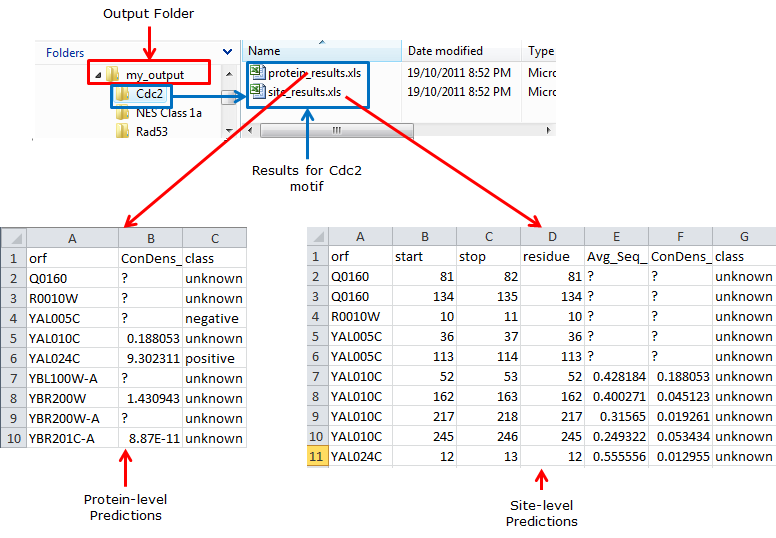
Figure 1: Structure of ConDens Output. In this example, the output data is piped to a folder called |
There are 7 columns in each
- orf: Name of the gene or protein.
- start: Start coordinate of motif.
- end: End coordinate of motif.
- residue: Coordinate for residue of interest (i.e. phosphorylated residue).
- Avg_Seq_Identity: Average sequence identity in local region around motif.
- ConDens_Score: Prediction score for this motif.
- label: Annotation for this moti. unknown denotes no information, positive denotes known substrate, and negative denotes known non-substrate
There are less columns in
Comments:
site_results.xls andprotein_results.xls are not excel files, although this author likes to use.xls file extension so that he can open the files as a spreadsheet with a double-click. Users who prefer other file extensions can choose to use.txt or.tab instead. See Guide to ConDens Predictor.site_results.[xls, txt, or tab] is the output file that is read by the ConDens Browser.- It is up to the user's discretion to organize the output data. In general, it is best to group output data by species and the set of multiple sequence alignments being used.